import matplotlib.pyplot as plt
import pandas as pd
import json
import numpy as np
from typing import Tuple
import matplotlib.pyplot as plt
# Set the desired RC parameters
rc_params = {
"xtick.direction": "in", # Mirrored ticks (in and out)
"ytick.direction": "in",
"xtick.top": True, # Show ticks on the top spine
"ytick.right": True, # Show ticks on the right spine
"xtick.major.size": 6, # Length of major ticks
"ytick.major.size": 6,
"xtick.minor.size": 4, # Length of minor ticks
"ytick.minor.size": 4,
"xtick.major.pad": 4, # Padding between tick and label
"ytick.major.pad": 4,
"xtick.minor.pad": 4,
"ytick.minor.pad": 4,
"font.size": 14, # Overall font size
"axes.labelsize": 16, # Font size of axis labels
"axes.titlesize": 18, # Font size of plot title
"xtick.labelsize": 12, # Font size of x-axis tick labels
"ytick.labelsize": 12, # Font size of y-axis tick labels
"xtick.major.width": 2, # Thickness of major ticks
"ytick.major.width": 2, # Thickness of major ticks
"xtick.minor.width": 1, # Thickness of minor ticks
"ytick.minor.width": 1, # Thickness of minor ticks
"lines.linewidth": 3, # Default linewidth for lines in plots
"patch.linewidth": 4, # Default linewidth for patches (e.g., rectangles, circles)
"axes.linewidth": 2, # Default linewidth for the axes spines
}
# Apply the RC parameters globally
plt.rcParams.update(rc_params)
JAX_LOGO = "https://github.com/jax-ml/jax/blob/main/images/jax_logo.png"
NUMPY_LOGO = "https://github.com/numpy/numpy/blob/main/branding/logo/primary/numpylogo.png"
CUPY_LOGO = (
"https://github.com/cupy/cupy/blob/main/docs/image/cupy_logo_1000px.png"
)
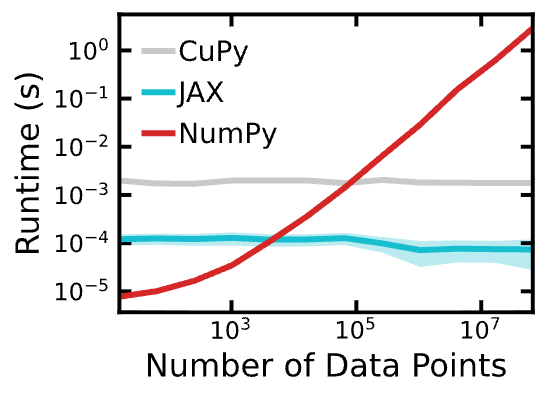
def plot_all_runtimes(cache_fn: str = "runtimes.json"):
fig, ax = plt.subplots(figsize=(4, 3.5))
with open(cache_fn, "r") as f:
data = json.load(f)
for label, runtimes in data.items():
runtimes = pd.DataFrame(runtimes)
_, _, line = plot(runtimes, ax=ax, label=label)
ax.legend(frameon=False)
return fig, ax
def plot(
runtimes: pd.DataFrame, ax=None, **kwgs
) -> Tuple[plt.Figure, plt.Axes]:
if ax is None:
fig, ax = plt.subplots(figsize=(4, 3.5))
fig = ax.figure
runtimes = runtimes.dropna()
runtimes = runtimes.sort_values(by="ND")
nds = runtimes["ND"].values
times, stds = runtimes["median"], runtimes["std"]
line = ax.plot(nds, times, **kwgs)
kwgs["label"] = None
ax.fill_between(
nds,
np.array(times) - np.array(stds),
np.array(times) + np.array(stds),
alpha=0.3,
**kwgs,
)
ax.set_yscale("log")
ax.set_xscale("log")
ax.set_xlabel("Number of Data Points")
ax.set_ylabel("Runtime (s)")
return fig, ax, line
fig, ax = plot_all_runtimes()
fig.savefig("runtimes.png", bbox_inches="tight")